doping{ }¶
- Calling sequence
structure{ region{ doping{ } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
items: \(\mathrm{maximum\;1}\)
- Functionality
Assigns dopants defined in the global group impurities{ } with selected concentrations.
- Examples
structure{ region{ doping{...} ... } } impurities{ ... }
Note
See — FREE — Schrödinger-Poisson - A comparison to the tutorial file of Greg Snider’s code as an example of use of gaussian1D{ } and gaussian2D{ } for donors and acceptors.
The Figure 2.5.1 shows a 3D doping profile that is defined inside a 20 nm x 20 nm x 50 nm cube where the 50 nm are the z direction. The doping rate profile is homogeneous with respect to the (x,y) plane, it only varies along the z direction.
The doping rate profile is constant between z = 10 nm and z = 25 nm with a rate of 1 x 1018 \([1/cm^3]\). It has Gaussian shape from z = 25 nm to z = 45 nm (gaussian1D). It is zero between z = 0 nm and z = 10 nm, as well as between z = 45 nm and z = 50 nm.
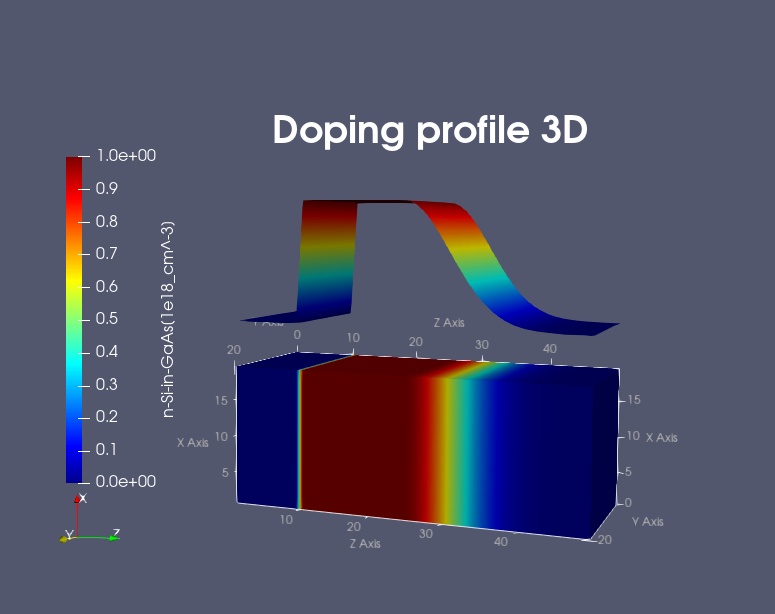
Figure 2.5.1 Three-dimensional doping profile (image generated by Exporting to ParaView).¶
position along z-direction (nm) |
generation rate (1/cm^3) |
|---|---|
0 ~ 10 nm |
0.0 |
10 ~ 25 nm |
constant (1.0 \(\times\) 1018) |
25 ~ 45 nm |
Gaussian (center = 25 nm, sigma_z = 6.0 nm) |
45 ~ 50 nm |
0.0 |
structure{ region{ everywhere{} binary{ name = GaAs } contact{ name = contact } } region{ binary{ name = GaAs } cuboid{ x = [0E0, 20E0] y = [0E0, 20E0] z = [0E0, 10E0] } } region{ binary{ name = GaAs } cuboid{ x = [0E0, 20E0] y = [0E0, 20E0] z = [10E0, 25E0] } doping{ constant{ name = "n-Si-in-GaAs" conc = 1.0E18 } } } region{ binary{ name = GaAs } cuboid{ x = [0E0, 20E0] y = [0E0, 20E0] z = [25E0, 45E0] } doping{ gaussian1D{ name = "n-Si-in-GaAs" conc = 1.0E18 z = 25 sigma_z = 6.0 } } } output_impurities{} } impurities{ donor{ name = "n-Si-in-GaAs" ... } } global{ simulate3D{} ... }
Nested keywords
remove{ }¶
- Calling sequence
structure{ region{ doping{ remove{ } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
items: \(\mathrm{no\;constraints}\)
- Functionality
Removes all dopants from a specific region.
- Example
structure{ region{ doping{ remove{} } ... } } impurities{ ... }
constant{ }¶
- Calling sequence
structure{ region{ doping{ constant{ } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
items: \(\mathrm{no\;constraints}\)
- Functionality
Defines constant doping profile over the region.
- Example
structure{ region{ doping{ constant{ name = "n-Si" conc = 1.0e18 add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } }
constant{ name }¶
- Calling sequence
structure{ region{ doping{ constant{ name } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{character\;string}\)
- Functionality
Refers to a dopant definition in impurities{ }.
- Example
structure{ region{ doping{ constant{ name = "n-Si" ... } } ... } } impurities{ donor{ name = "n-Si" ... } }
constant{ conc }¶
- Calling sequence
structure{ region{ doping{ constant{ conc } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{cm^{-3}}\)
- Functionality
Defines value dopant concentration.
- Example
structure{ region{ doping{ constant{ conc = 1.0e18 ... } } ... } } impurities{ ... }
constant{ add }¶
- Calling sequence
structure{ region{ doping{ constant{ add } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
type: \(\mathrm{choice}\)
choices:
yes;nodefault:
yes
- Functionality
Choses the mode of assigning doping. If
add = yesthen the doping in the region is added to already defined ones. Otherwise, the previously defined doping is replaced.- Example
structure{ region{ doping{ constant{ add = no ... } } ... } } impurities{ ... }
linear{ }¶
- Calling sequence
structure{ region{ doping{ linear{ } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
items: \(\mathrm{no\;constraints}\)
- Functionality
Defines linear doping profile along a defined line
- Examples
structure{ region{ doping{ linear{ name = "n-Si" conc = [ 1.0e18, 2.0e18 ] x = [ 50.0, 100.0 ] add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } } global{ simulate1D{ } }
structure{ region{ doping{ linear{ name = "n-Si" conc = [ 1.0e18, 2.0e18 ] x = [ 50.0, 100.0 ] y = [ 50.0, 100.0 ] z = [ 50.0, 100.0 ] add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } } global{ simulate3D{ } }
linear{ name }¶
- Calling sequence
structure{ region{ doping{ linear{ name } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{character\;string}\)
- Functionality
Refers to a dopant definition in impurities{ }.
- Example
structure{ region{ doping{ linear{ name = "n-Si" ... } } ... } } impurities{ donor{ name = "n-Si" ... } }
linear{ conc }¶
- Calling sequence
structure{ region{ doping{ linear{ conc } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{vector\;of\;2\;real\;numbers}\)
values:
[0.0, ...)for every dimensionunit: \(\mathrm{cm^{-3}}\)
- Functionality
Defines values of linear dopant profile at the ends of the ends of defined line. The first value corresponds to the starting point of the line and the second value to the ending point of the line.
- Example
structure{ region{ doping{ linear{ conc = [ 1.0e18, 2.0e18 ] ... } } ... } } impurities{ ... }
linear{ x }¶
- Calling sequence
structure{ region{ doping{ linear{ x } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{vector\;of\;2\;real\;numbers}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines x-coordinates of the starting point and ending point of the line, along which the linear distribution of dopants is defined.
- Example
structure{ region{ doping{ linear{ x = [ 50.0, 100.0 ] ... } } ... } } impurities{ ... }
linear{ y }¶
- Calling sequence
structure{ region{ doping{ linear{ y } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{vector\;of\;2\;real\;numbers}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines y-coordinates of the starting point and ending point of the line, along which the linear distribution of dopants is defined.
- Example
structure{ region{ doping{ linear{ x = [ 50.0, 100.0 ] y = [ 50.0, 100.0 ] ... } } ... } } impurities{ ... }
linear{ z }¶
- Calling sequence
structure{ region{ doping{ linear{ z } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{vector\;of\;2\;real\;numbers}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines z-coordinates of the starting point and ending point of the line, along which the linear distribution of dopants is defined.
- Example
structure{ region{ doping{ linear{ x = [ 50.0, 100.0 ] y = [ 50.0, 100.0 ] z = [ 50.0, 100.0 ] ... } } ... } } impurities{ ... }
linear{ add }¶
- Calling sequence
structure{ region{ doping{ linear{ add } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
type: \(\mathrm{choice}\)
choices:
yes;nodefault:
yes
- Functionality
Choses the mode of assigning doping. If
add = yesthen the doping in the region is added to already defined ones. Otherwise, the previously defined doping is replaced.- Example
structure{ region{ doping{ linear{ add = no ... } } ... } } impurities{ ... }
gaussian1D{ }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
items: \(\mathrm{no\;constraints}\)
- Functionality
Defines Gaussian distribution function in one direction, constant in remaining perpendicular directions.
Note
This profile corresponds to LSS theory (Lindhard, Scharff, Schiott theory) for doping - Gaussian distribution of ion implantation.
- Examples
structure{ region{ doping{ gaussian1D{ name = "n-Si" conc = 1.0e18 x = 50.0 sigma_x = 5.0 add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } } global{ simulate1D{ } }
structure{ region{ doping{ gaussian1D{ name = "n-Si" dose = 1e12 y = 50.0 sigma_y = 5.0 add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } } global{ simulate2D{ } }
gaussian1D{ name }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ name } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{character\;string}\)
- Functionality
Refers to a dopant definition in impurities{ }.
- Example
structure{ region{ doping{ gaussian1D{ name = "n-Si" ... } } ... } } impurities{ donor{ name = "n-Si" ... } }
gaussian1D{ conc }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ conc } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{cm^{-3}}\)
- Functionality
Defines maximum of dopant concentration.
- Example
structure{ region{ doping{ gaussian1D{ conc = 1.0e18 ... } } ... } } impurities{ ... }
gaussian1D{ dose }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ dose } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{cm^{-2}}\)
- Functionality
Defines implantation dose. It is an integrated density of Gaussian function conc = dose / ( SQRT(2*pi) * sigma_x ).
Hint
Typical values range from 1e11 cm-2 to 1e16 cm-2.
- Example
structure{ region{ doping{ gaussian1D{ dose = 1.0e12 ... } } ... } } impurities{ ... }
gaussian1D{ x }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ x } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the x-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian1D{ x = 50.0 ... } } ... } } impurities{ ... }
gaussian1D{ y }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ y } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the y-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian1D{ y = 50.0 ... } } ... } } impurities{ ... }
gaussian1D{ z }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ z } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the z-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian1D{ z = 50.0 ... } } ... } } impurities{ ... }
gaussian1D{ sigma_x }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ sigma_x } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the x-axis.
- Example
structure{ region{ doping{ gaussian1D{ x = 50.0 sigma_x = 5.0 ... } } ... } } impurities{ ... }
gaussian1D{ sigma_y }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ sigma_y } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the y-axis.
- Example
structure{ region{ doping{ gaussian1D{ y = 50.0 sigma_y = 5.0 ... } } ... } } impurities{ ... }
gaussian1D{ sigma_z }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ sigma_z } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the z-axis.
- Example
structure{ region{ doping{ gaussian1D{ z = 50.0 sigma_z = 5.0 ... } } ... } } impurities{ ... }
gaussian1D{ add }¶
- Calling sequence
structure{ region{ doping{ gaussian1D{ add } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
type: \(\mathrm{choice}\)
choices:
yes;nodefault:
yes
- Functionality
Choses the mode of assigning doping. If
add = yesthen the doping in the region is added to already defined ones. Otherwise, the previously defined doping is replaced.- Example
structure{ region{ doping{ gaussian1D{ add = no ... } } ... } } impurities{ ... }
gaussian2D{ }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
items: \(\mathrm{no\;constraints}\)
- Functionality
Defines Gaussian distribution function in two directions, constant in remaining perpendicular direction.
- Examples
structure{ region{ doping{ gaussian2D{ name = "n-Si" conc = 1.0e18 x = 50.0 y = 50.0 sigma_x = 5.0 sigma_y = 5.0 add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } } global{ simulate2D{ } }
structure{ region{ doping{ gaussian2D{ name = "n-Si" dose = 1e6 x = 50.0 z = 50.0 sigma_x = 5.0 sigma_z = 5.0 add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } } global{ simulate3D{ } }
gaussian2D{ name }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ name } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{character\;string}\)
- Functionality
Refers to a dopant definition in impurities{ }.
- Example
structure{ region{ doping{ gaussian2D{ name = "n-Si" ... } } ... } } impurities{ donor{ name = "n-Si" ... } }
gaussian2D{ conc }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ conc } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{cm^{-3}}\)
- Functionality
Defines maximum of dopant concentration.
- Example
structure{ region{ doping{ gaussian2D{ conc = 1.0e18 ... } } ... } } impurities{ ... }
gaussian2D{ dose }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ dose } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{cm^{-1}}\)
- Functionality
Defines implantation dose. It is an integrated density of Gaussian function.
- Example
structure{ region{ doping{ gaussian2D{ dose = 1.0e6 ... } } ... } } impurities{ ... }
gaussian2D{ x }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ x } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the x-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian2D{ x = 50.0 y = 50.0 ... } } ... } } impurities{ ... }
gaussian2D{ y }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ y } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the y-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian2D{ y = 50.0 z = 50.0 ... } } ... } } impurities{ ... }
gaussian2D{ z }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ z } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the z-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian2D{ z = 50.0 x = 50.0 ... } } ... } } impurities{ ... }
gaussian2D{ sigma_x }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ sigma_x } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the x-axis.
- Example
structure{ region{ doping{ gaussian2D{ x = 50.0 y = 50.0 sigma_x = 5.0 sigma_y = 5.0 ... } } ... } } impurities{ ... }
gaussian2D{ sigma_y }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ sigma_y } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the y-axis.
- Example
structure{ region{ doping{ gaussian2D{ y = 50.0 z = 50.0 sigma_y = 5.0 sigma_z = 5.0 ... } } ... } } impurities{ ... }
gaussian2D{ sigma_z }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ sigma_z } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the z-axis.
- Example
structure{ region{ doping{ gaussian2D{ z = 50.0 x = 50.0 sigma_z = 5.0 sigma_x = 5.0 ... } } ... } } impurities{ ... }
gaussian2D{ add }¶
- Calling sequence
structure{ region{ doping{ gaussian2D{ add } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
type: \(\mathrm{choice}\)
choices:
yes;nodefault:
yes
- Functionality
Choses the mode of assigning doping. If
add = yesthen the doping in the region is added to already defined ones. Otherwise, the previously defined doping is replaced.- Example
structure{ region{ doping{ gaussian2D{ add = no ... } } ... } } impurities{ ... }
gaussian3D{ }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
items: \(\mathrm{no\;constraints}\)
- Functionality
Defines Gaussian distribution function in three directions.
- Example
structure{ region{ doping{ gaussian3D{ name = "n-Si" conc = 1.0e18 x = 50.0 y = 50.0 z = 50.0 sigma_x = 5.0 sigma_y = 5.0 sigma_z = 5.0 add = no } } ... } } impurities{ donor{ name = "n-Si" degeneracy = 2 energy = 0.0058 } } global{ simulate3D{ } }
gaussian3D{ name }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ name } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{character\;string}\)
- Functionality
Refers to a dopant definition in impurities{ }.
- Example
structure{ region{ doping{ gaussian3D{ name = "n-Si" ... } } ... } } impurities{ donor{ name = "n-Si" ... } }
gaussian3D{ conc }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ conc } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{cm^{-3}}\)
- Functionality
Defines maximum of dopant concentration.
- Example
structure{ region{ doping{ gaussian3D{ conc = 1.0e18 ... } } ... } } impurities{ ... }
gaussian3D{ dose }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ dose } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{-}\)
- Functionality
Defines implantation dose. It is an integrated density of Gaussian function.
- Example
structure{ region{ doping{ gaussian3D{ dose = 1.0 ... } } ... } } impurities{ ... }
gaussian3D{ x }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ x } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the x-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian3D{ x = 50.0 y = 50.0 z = 50.0 ... } } ... } } impurities{ ... }
gaussian3D{ y }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ y } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the y-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian3D{ x = 50.0 y = 50.0 z = 50.0 ... } } ... } } impurities{ ... }
gaussian3D{ z }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ z } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values: no constraints
unit: \(\mathrm{nm}\)
- Functionality
Defines the z-coordinate of the center of the Gauss distribution.
- Example
structure{ region{ doping{ gaussian3D{ x = 50.0 y = 50.0 z = 50.0 ... } } ... } } impurities{ ... }
gaussian3D{ sigma_x }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ sigma_x } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the x-axis.
- Example
structure{ region{ doping{ gaussian3D{ x = 50.0 y = 50.0 z = 50.0 sigma_x = 5.0 sigma_y = 5.0 sigma_z = 5.0 ... } } ... } } impurities{ ... }
gaussian3D{ sigma_y }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ sigma_y } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the y-axis.
- Example
structure{ region{ doping{ gaussian3D{ x = 50.0 y = 50.0 z = 50.0 sigma_x = 5.0 sigma_y = 5.0 sigma_z = 5.0 ... } } ... } } impurities{ ... }
gaussian3D{ sigma_z }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ sigma_z } } } }- Properties
usage: \(\mathrm{\textcolor{Dandelion}{conditional}}\)
type: \(\mathrm{real\;number}\)
values:
[0.0, ...)unit: \(\mathrm{nm}\)
- Functionality
Defines standard deviation of the Gauss distribution along the z-axis.
- Example
structure{ region{ doping{ gaussian3D{ x = 50.0 y = 50.0 z = 50.0 sigma_x = 5.0 sigma_y = 5.0 sigma_z = 5.0 ... } } ... } } impurities{ ... }
gaussian3D{ add }¶
- Calling sequence
structure{ region{ doping{ gaussian3D{ add } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
type: \(\mathrm{choice}\)
choices:
yes;nodefault:
yes
- Functionality
Choses the mode of assigning doping. If
add = yesthen the doping in the region is added to already defined ones. Otherwise, the previously defined doping is replaced.- Example
structure{ region{ doping{ gaussian3D{ add = no ... } } ... } } impurities{ ... }
import{ }¶
- Calling sequence
structure{ region{ doping{ import{ } } } }- Properties
usage: \(\mathrm{\textcolor{ForestGreen}{optional\;within\;the\;group}}\)
items: \(\mathrm{no\;constraints}\)
- Functionality
Imports generation profile from an external file and adds it to already defined (if defined) profiles.
- Example
structure{ region{ doping{ import{ name = "n-Si" import_from = "importing_dopant_profile" } } ... } } impurities{ donor{ name = "n-Si" ... } } import{ file{ name = "importing_dopant_profile" filename = "precious_dopant_profile.dat" ... } }
import{ name }¶
- Calling sequence
structure{ region{ doping{ import{ name } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{character\;string}\)
- Functionality
Refers to a dopant definition in impurities{ }.
- Example
structure{ region{ doping{ import{ name = "n-Si" ... } } ... } } impurities{ donor{ name = "n-Si" ... } } import{ ... }
import{ import_from }¶
- Calling sequence
structure{ region{ doping{ import{ import_from } } } }- Properties
usage: \(\mathrm{\textcolor{WildStrawberry}{required\;within\;the\;group}}\)
type: \(\mathrm{character\;string}\)
- Functionality
Reference to imported data in import{ }.
- Example
structure{ region{ doping{ import{ import_from = "importing_dopant_profile" ... } } ... } } impurities{ ... } import{ file{ name = "importing_dopant_profile" filename = "precious_dopant_profile.dat" ... } }