— NEW — Importing files¶
Header¶
- Files for the tutorial located in nextnano++\examples\basics
import-dat_1D_nnp.in - importing *.dat files to 1D simulation
import-dat_2D_nnp.in - importing *.dat files to 2D simulation
import-dat_3D_nnp.in - importing *.dat files to 3D simulation
- Scope of the tutorial:
This tutorial is presenting how to import various files to nextnano++ simulations. The examples cover importing electric potential, alloy contents, and strain for 1D, 2D, and 3D simulations from *.dat files.
- Relevant output Files:
bias_00000\bandedges.dat
Imports\Ternary_alloy.dat
Imports\Strain_Tensor.dat
Imports\Potential.dat
- Introduced Keywords:
import{ directory file{ filename format } }region{ ternary_import{ } }strain{ import_strain{ } }poisson{ import_potential{} }
Importing data¶
Reading external files¶
The pivotal group responsible for importing files for simulations with nextnano++ is the group import{ }. Its purpose for this tutorial is to inform nextnano++ about:
the location of a selected file,
format of the file,
name of the file,
how to refer to the file,
whether the data should be rescaled.
- EXAMPLE 1. Importing a file from the location of the input file
Let us say that one has an input file
C:\input_files\my_input_file.in. Having the following script in the fileimport{ file{ name = "some_imported_data" filename = "my_alloy_from_XRF" format = DAT } }results in nextnano++ trying to access and read a file
C:\input_files\my_alloy_from_XRF.dat. The file can be used in the input file under the namesome_imported_data.- EXAMPLE 2. Importing a file from an arbitrary location
Giving that the data to import is stored elsewhere, one just needs to define
import{ directory }attribute to navigate nextnano++ to the location of the file to import. The scriptimport{ directory = "D:\\my_precious_measurements\\" file{ name = "some_imported_data" filename = "my_alloy_from_XRF" format = DAT } }instructs nextnano++ to access and read
D:\my_precious_measurements\my_alloy_from_XRF.dat.Hint
It is also allowed to write
directory = "D:\my_precious_measurements\"instead ofdirectory = "D:\\my_precious_measurements\\"Note
The input files prepared for this tutorial have
import{ directory = "./"}specified which is equivalent to not specifyingdirectoryattribute at all.- EXAMPLE 3. Importing a file and rescaling
Let us assume that somebody has prepared a file containing alloy content defined in the range from 0 to 100. The nextnano++ tool requires the content to be imported as mole fraction, therefore, defined in the range from 0 to 1. To import data from such a file one can use a scaling factor
0.01which will be used while reading the file. Running the following scriptimport{ directory = "D:\\my_precious_measurements\\" file{ name = "some_imported_data" filename = "my_alloy_from_XRF" format = DAT scale = 0.01 } }results in nextnano++ rescaling all the imported values (except the domain, coordinates) by multiplying them by
0.01. Therefore, a data for 2D simulation
coord-x
coord-y
alloy-x
0
3
10
5
5
25
20
6
70
will be read as
coord-x
coord-y
alloy-x
0
3
0.1
5
5
0.25
20
6
0.7
To use imported file in the simulation, one needs to use the reference name specified by import{ file{ name } } in other proper places in the input file.
Electric potential¶
For this tutorial we provide you with three files containing electric potential for importing import-dat_1D_nnp_potential.dat, import-dat_2D_nnp_potential.dat*, and import-dat_3D_nnp_potential.dat for 1D, 2D, and 3D simulations, respectively. Let us consider 1D simulation for simplicity; 2D and 3D cases are similar.
- EXAMPLE 4. Importing electric potential from a
*.datfileThe :import_1D_dat_nnp_potential.dat is imported in the input file :import-dat_1D_nnp.in as follows.
import{ file{ name = "imported_potential" filename = "import-dat_1D_nnp_potential.dat" format = DAT } }It allows nextnano++ to use the data through the name
"imported_potential"elsewhere. As the electric potential is related to the Poisson equation, one needs to use the name inside a nested grouppoisson{ import{ } }in order to inform the tool that these data should be used as an electric potential. The relevant piece of script in the :import_1D_dat_nnp.in is:poisson{ import_potential{ import_from = "imported_potential" } }
Strain tensor¶
You can apply these manners to the other parameters, such as, strain and potential.
100import{
101
102 file{
103 name = "imported_strain" # name for referencing the imported data in the input file
104 filename = "import-dat_1D_nnp_strain.dat" # name of file which is imported
105 format = DAT # format of the file to be imported. At the moment only AVS format and a simple .dat format is supported.
106 }
107}
77strain{
78 import_strain{
79 import_from = "imported_strain" # reference to imported data in import{ }. The file being imported must have exactly six data components
80 # expected order of tensor components is: e_11, e_22, e_33, e_12, e_13, e_23.
81 }
82 output_strain_tensor{
83 simulation_system = yes
84 crystal_system = yes
85 }
86}
In the case, the import file has only one column (x) of a coordinate. Number of required columns of coordinate depends on dimensionality of the simulation, 2 columns (x and y) are necessary for 2D simulation and 3 columns (x, y, and z) for 3D simulation. Additionally, the file contains 6 tensor components, \(\varepsilon_{\mathrm{11}}\), \(\varepsilon_{\mathrm{22}}\), \(\varepsilon_{\mathrm{33}}\), \(\varepsilon_{\mathrm{12}}\), \(\varepsilon_{\mathrm{13}}\), and \(\varepsilon_{\mathrm{23}}\), each in separate collumn.
Alloy compositions¶
24structure{
25 output_alloy_composition{}
26 region{
27 line{
28 x = [0, 16]
29 }
30 ternary_import{
31 name = "Al(x)Ga(1-x)As" # ternary material name for this region which uses imported alloy profile
32 import_from = "imported_ternary" # reference to imported data in import{ }. The file being imported must have exactly one data component (x)
33 }
34 }
35}
As ternary_import{ } is used to import alloy profile, imported_ternary file contains information about alloy profile.
The file has the following data.
x-coord |
alloy_parameter |
|---|---|
4 |
15 |
12 |
30 |
The “alloy_parameter” should be \(\le\) 1, therefore, import{ file{ scale } } is necessary to be consistent with that.
Once you import a file, you can use it multiple times.
100import{
101
102 file{
103 name = "imported_quaternary" # name for referencing the imported data in the input file
104 filename = "import-dat_1D_nnp_quaternary.dat" # name of file which is imported
105 format = DAT # format of the file to be imported.
106 # At the moment only AVS format and a simple .dat format is supported.
107 }
108}
24structure{
25
26 region{
27 line{
28 x = [17, 33]
29 }
30 quaternary_import{
31 name = "Al(x)Ga(y)In(1-x-y)As" # quaternary material name for this region which uses imported alloy profile
32 import_from = "imported_quaternary" # reference to imported data in import{ }.
33 # sThe file being imported must have exactly two data components (x,y).
34 }
35 }
36 region{
37 line{
38 x = [34, 50]
39 }
40 quaternary_import{
41 name = "Al(x)Ga(1-x)As(y)Sb(1-y)" # quaternary material name for this region which uses imported alloy profile
42 import_from = "imported_quaternary" # reference to imported data in import{ }.
43 # The file being imported must have exactly two data components (x,y).
44 }
45 }
46}
In the code, you are using import-dat_1D_nnp_quaternary.dat file twice to specify those alloy compositions.
Imported data in the simulation¶
import{ output_imports{} } outputs all imported data including scale factors. The filenames of the outputs correspond to the ones defined import{ file{ name } }.
Electric potential¶
Attention
Prepared input files are not solving the Poisson equation.
The Figure 2.4.23 shows the potential defined in the import files.
Figure 2.4.23 The potential introduced from the import file. The resulting potential in the entire structure.¶
Strain tensor¶
Figure 2.4.24 Imported strain tensor.¶
Alloy compositions¶
Figure 2.4.25 shows the alloy compositions in each region defined in the import files (a), (b) and the input file (c).
Figure 2.4.25 The alloy composition of Al(x)Ga(1-x)As is shown in (a). The alloy composition of Al(x)Ga(y)In(1-x-y)As is shown in (b) (The violet line: x, The purple line: y). (c) shows the alloy compositions in the whole structure. Region I is Al(x)Ga(y)In(1-x-y)As, region II is Al(x)Ga(y)In(1-x-y)As, and region III is Al(x)Ga(1-x)As(y)Sb(1-y), respectively.¶
The grid points in Figure 2.4.25 are originated from the import files.
There are some important points you can see from Figure 2.4.25 (c). First, you should be aware that the values between grid points are interpolated linearly. Therefore, the composition between the region I and the region II steeply drops. Second, the regions in which any date is not specified in import files are interpolated by constants. As the composition of the region III is not specified in the import files, it has continuously taken over the value at the boundary between the region II and the region III.
Resulting bandedges¶
At last, we briefly check the band edges of the structure (Figure 2.4.26).
Figure 2.4.26 The band edges of the structure. The HH band and the LH band are degenerated in the region where there is no strain.¶
HH, LH, and SO band mean heavy hole, light hole, and split off band, respectively. The Fermi level is set to \(0~\mathrm{eV}\) You can refer to Definition of Band Offsets (zincblende) for further knowledge about band offsets.
2D and 3D simulations¶
In the 2D simulation, you can import files in the same manners as in the 1D simulation. Of course, the import files have to be 2 dimensional.
Figure 2.4.27 shows the geometry of the materials used in this simulation.
Figure 2.4.27 The geometry of the materials used in the 2D simulation. The dashed line is along x = 7.5 nm.¶
Here, we look at the alloy compositions of the materials as an example of a 2D import file. 2D_ternary_alloy.dat, 2D_quaternary_alloy.dat are imported for specifying the alloy compositions for the materials above.
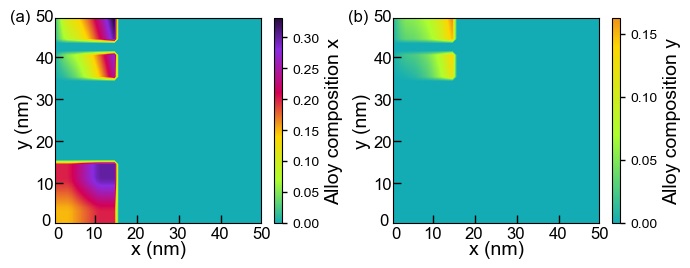
Figure 2.4.28 The alloy compositions of (a) x and (b) y of the materials used in the 2D simulation.¶
Attention
In this tutorial we are assuming always that the imported data is defined on a domain or subspace of the simulation domain. Therefore, the number of dimensions of the domain of the imported data is always assumed to be the same as of the simulation, e.g., 2D simulation imports data with two first columns standing for x and y coordinates. If you need a tutorial covering such case, let us know here.
Last update: 16/07/2024